In a current examine revealed within the journal Nature Machine Intelligence, researchers developed “DeepGO-SE,” a technique to foretell gene ontology (GO) features from protein sequences utilizing a big, pre-trained protein language mannequin.
 Research: Protein operate prediction as approximate semantic entailment. Picture Credit score: DarwinAmelie / Shutterstock
Research: Protein operate prediction as approximate semantic entailment. Picture Credit score: DarwinAmelie / Shutterstock
Though protein construction prediction has more and more change into correct through the years, protein operate prediction is difficult because of the restricted variety of identified features, compounded by their interactions and complexity. GOs are used to explain protein features. GO consists of three sub-ontologies describing molecular features (MFO) of proteins, their function in organic processes (BPO), and mobile parts (CCO) the place they’re energetic.
A big limitation of a number of operate prediction strategies is their reliance on sequence similarity. Though efficient for proteins with comparable sequences and well-characterized features, this method is much less dependable for these with no or little sequence similarity. Furthermore, protein features are based on their construction, and proteins with comparable buildings might have dissimilar sequences.
The background information contained in axioms of GOs will be leveraged by way of machine studying fashions for improved predictions. There are only some strategies that make the most of the formal axioms in GOs. Hierarchical classification strategies, comparable to DeePred, TALE, DeepGO, and GOStruct2 use subsumption axioms however ignore others that might be used to restrict search house and improve predictions.
The examine and findings
Within the current examine, researchers developed a protein operate prediction technique, DeepGO-SE, utilizing a big, pre-trained protein language mannequin. DeepGO-SE applied knowledge-enhanced studying by way of semantic entailment in three steps. First, an approximate mannequin was generated utilizing ELEmbeddings primarily based on logical concept consisting of GO axioms (background information) and assertions about proteins like “protein has a operate C.”
Subsequent, single proteins have been represented by evolutionary scale mannequin 2 (ESM2) embeddings and used as situations within the approximate mannequin to maximise the assertion’s fact as an optimization goal. Lastly, this process was repeated to generate ok approximate fashions; entailment was outlined as the reality in all fashions, and the ok fashions have been utilized for approximate semantic entailment.
The researchers in contrast their technique with 5 baseline strategies utilizing a UniProtKB/Swiss-Prot dataset. Baseline strategies have been naïve method, multilayer perceptron (MLP), DeepGraphGO, DeepGoZero, and DeepGOCNN. GO sub-ontologies have been individually skilled and evaluated. DeepGO-SE considerably outperformed the baseline strategies.
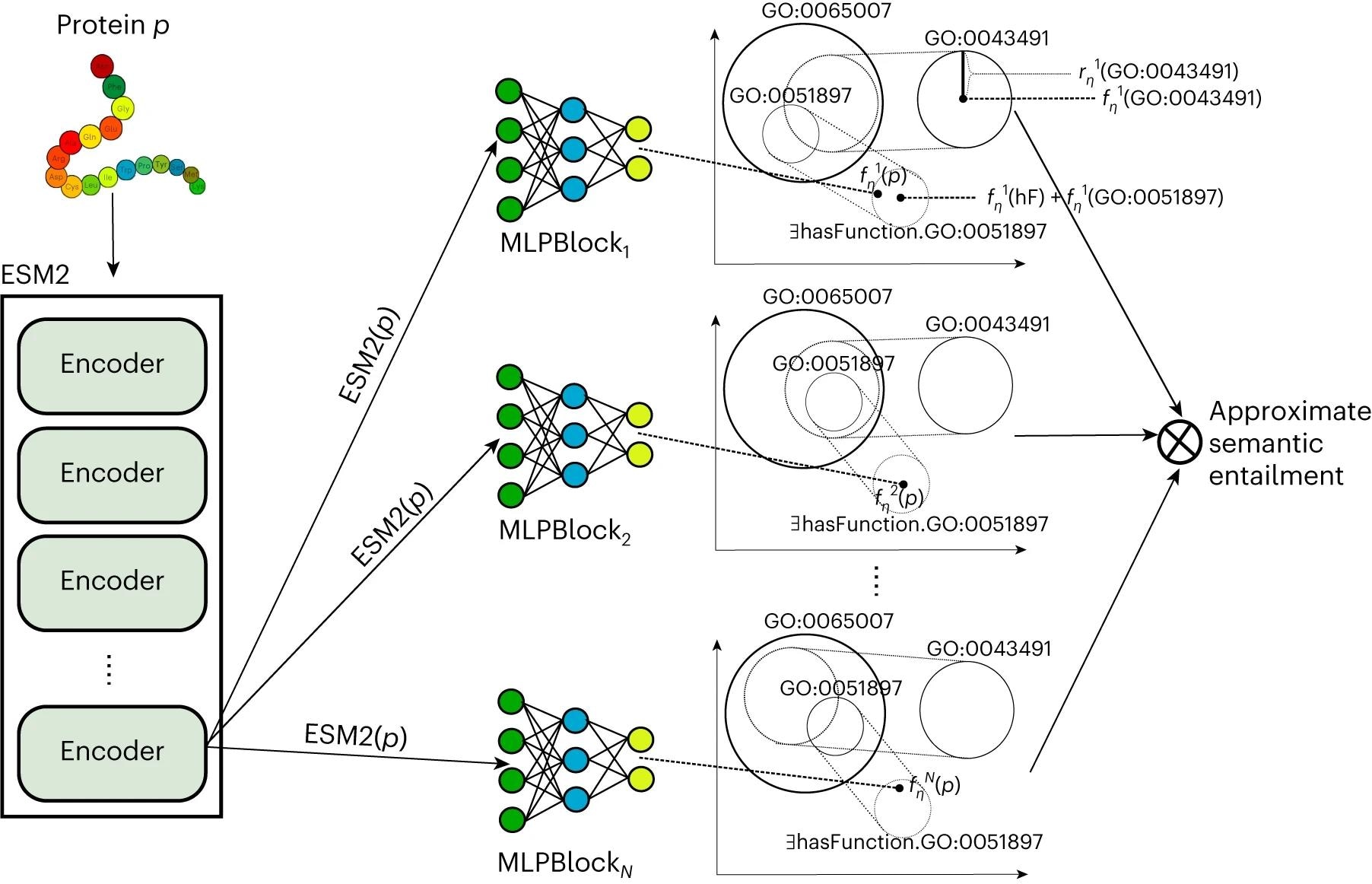
Left: protein p is embedded in a vector house utilizing ESM2 mannequin. Proper: a number of fashions with an MLP that embeds the protein in the identical house because the GO axioms. Moreover, predictions from a number of fashions are mixed to carry out approximate semantic entailment.
In MFO, the utmost F measure (F max) of DeepGO-SE was 0.554, 7% bigger than that of DeepGoZero and MLP strategies. In BPO, its F max (0.432) was 8% increased than DeepGraphGO. In CCO, DeepGO-SE achieved an F max of 0.721. Subsequent, the group modified the protein embeddings to encode extra data relating to the proteome and its interactions.
To this finish, enter vector(s) to DeepGO-SE have been altered, and three experiments have been carried out. First, ESM2 embeddings have been used as enter for every protein in DeepGOGAT-SE. Subsequent, experimental annotations of a protein to molecular features have been used as enter in DeepGOGATMF-SE. Lastly, DeepGO-SE model-derived prediction scores for molecular features have been used because the enter in DeepGOGATMF-SE-Pred.
Combining ESM2 embeddings and protein-protein interactions (PPIs) in DeepGOGAT-SE decreased the efficiency of MFO prediction (F max: 0.525) however marginally improved the minimal semantic distance (S min). Moreover, BPO prediction was improved (F max: 0.435). Notably, the perfect BPO efficiency was noticed with DeepGOGATMF-SE (F max: 0.448), adopted by DeepGOGATMF-SE-Pred (F max: 0.444). Integrating PPIs in DeepGO-SE elevated the F max for CCOs to 0.736.
The group additionally evaluated their baseline strategies utilizing the neXtPro dataset (of manually predicted protein features). They discovered that DeepGO-SE achieved the perfect F max (0.386). DeepGOGAT-SE carried out the perfect for BPOs, with an F max of 0.35. The group couldn’t consider the DeepGOGATMF-SE-Pred technique as a result of many proteins lacked handbook molecular features.
Lastly, an ablation examine was carried out to evaluate the contribution of particular person parts of the fashions. ELEmbeddings axiom loss features have been eliminated for every mannequin, and performance prediction loss was optimized. Eradicating axiom losses from DeepGO-SE lowered MFO efficiency with out impacting BPO and CCO efficiency.
In DeepGOGAT-SE, eradicating axioms and semantic entailment modules barely improved the efficiency of MFO however lowered that of BPO and CCO. BPO and CCO efficiency was higher when axioms and semantic entailment have been eliminated in fashions utilizing molecular features and PPIs as options.
Conclusions
Taken collectively, DeepGO-SE is an improved protein operate prediction technique that includes sequence options derived from a pre-trained protein language mannequin, GO background information, and PPIs. It may well predict BPO and CCO from a protein sequence alone; nevertheless, PPI data was required for greatest outcomes. As a result of many novel proteins lack identified interactions, strategies that predict interactions for novel proteins from their sequence solely are vital.
Journal reference:
- Kulmanov M, Guzmán-Vega FJ, Duek Roggli P, Lane L, Arold ST, Hoehndorf R. Protein operate prediction as approximate semantic entailment. Nat Mach Intell. Revealed on-line February 14, 2024, DOI: 10.1038/s42256-024-00795-w, https://www.nature.com/articles/s42256-024-00795-w


